Let’s use the mammal sleep dataset from ggplot2. This dataset
contains the number of hours spent sleeping per day for 83 different species of
mammals along with each species’ brain mass (kg) and body mass (kg), among other
measures. Here’s a first look at the data.
- library(dplyr, warn.conflicts = FALSE)
- library(ggplot2)
-
- # Preview sorted by brain/body ratio. I chose this sorting so that humans would
- # show up in the preview.
- msleep %>%
- select(name, sleep_total, brainwt, bodywt, everything()) %>%
- arrange(desc(brainwt / bodywt))
- #> # A tibble: 83 × 11
- #> name sleep_total brainwt bodywt genus
- #> <chr> <dbl> <dbl> <dbl> <chr>
- #> 1 Thirteen-lined ground squirrel 13.8 0.00400 0.101 Spermophilus
- #> 2 Owl monkey 17.0 0.01550 0.480 Aotus
- #> 3 Lesser short-tailed shrew 9.1 0.00014 0.005 Cryptotis
- #> 4 Squirrel monkey 9.6 0.02000 0.743 Saimiri
- #> 5 Macaque 10.1 0.17900 6.800 Macaca
- #> 6 Little brown bat 19.9 0.00025 0.010 Myotis
- #> 7 Galago 9.8 0.00500 0.200 Galago
- #> 8 Mole rat 10.6 0.00300 0.122 Spalax
- #> 9 Tree shrew 8.9 0.00250 0.104 Tupaia
- #> 10 Human 8.0 1.32000 62.000 Homo
- #> # ... with 73 more rows, and 6 more variables: vore <chr>, order <chr>,
- #> # conservation <chr>, sleep_rem <dbl>, sleep_cycle <dbl>, awake <dbl>
-
- ggplot(msleep) +
- aes(x = brainwt, y = sleep_total) +
- geom_point()
- #> Warning: Removed 27 rows containing missing values (geom_point).
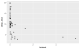
Hmmm, not very helpful! We should put our measures on a log-10 scale. Also, 27
of the species don’t have brain mass data, so we’ll exclude those rows for the
rest of this tutorial.
- msleep <- msleep %>%
- filter(!is.na(brainwt)) %>%
- mutate(log_brainwt = log10(brainwt),
- log_bodywt = log10(bodywt),
- log_sleep_total = log10(sleep_total))
Now, plot the log-transformed data. But let’s also get a little fancy and label
the points for some example critters :cat: so that we can get some intuition
about the data in this scaling. (Plus, I wanted to try out the annotation
tips from the R4DS book.)
- # Create a separate data-frame of species to highlight
- ex_mammals <- c("Domestic cat", "Human", "Dog", "Cow", "Rabbit",
- "Big brown bat", "House mouse", "Horse", "Golden hamster")
-
- # We will give some familiar species shorter names
- renaming_rules <- c(
- "Domestic cat" = "Cat",
- "Golden hamster" = "Hamster",
- "House mouse" = "Mouse")
-
- ex_points <- msleep %>%
- filter(name %in% ex_mammals) %>%
- mutate(name = stringr::str_replace_all(name, renaming_rules))
-
- # Define these labels only once for all the plots
- lab_lines <- list(
- brain_log = "Brain mass (kg., log-scaled)",
- sleep_raw = "Sleep per day (hours)",
- sleep_log = "Sleep per day (log-hours)"
- )
-
- ggplot(msleep) +
- aes(x = brainwt, y = sleep_total) +
- geom_point(color = "grey40") +
- # Circles around highlighted points + labels
- geom_point(size = 3, shape = 1, color = "grey40", data = ex_points) +
- ggrepel::geom_text_repel(aes(label = name), data = ex_points) +
- # Use log scaling on x-axis
- scale_x_log10(breaks = c(.001, .01, .1, 1)) +
- labs(x = lab_lines$brain_log, y = lab_lines$sleep_raw)

As a child growing up on a dairy farm :cow:, it was remarkable to me how little
I saw cows sleeping, compared to dogs or cats. Were they okay? Are they
constantly tired and groggy? Maybe they are asleep when I’m asleep? Here, it
looks like they just don’t need very much sleep.
Next, let’s fit a classical regression model. We will use a log-scaled sleep
measure so that the regression line doesn’t imply negative sleep (even though
brains never get that large).
- m1_classical <- lm(log_sleep_total ~ log_brainwt, data = msleep)
- arm::display(m1_classical)
- #> lm(formula = log_sleep_total ~ log_brainwt, data = msleep)
- #> coef.est coef.se
- #> (Intercept) 0.74 0.04
- #> log_brainwt -0.13 0.02
- #> ---
- #> n = 56, k = 2
- #> residual sd = 0.17, R-Squared = 0.40
We can interpret the model in the usual way: A mammal with 1 kg (0 log-kg)
of brain mass sleeps 100.74 = 5.5 hours per
day. A mammal with a tenth of that brain mass (-1 log-kg) sleeps
100.74 + 0.13 = 7.4 hours.
We illustrate the regression results to show the predicted mean of y and
its 95% confidence interval. This task is readily accomplished in ggplot2 using
stat_smooth(). This function fits a model and plots the mean and CI for each
aesthetic grouping of data1 in a plot.
- ggplot(msleep) +
- aes(x = log_brainwt, y = log_sleep_total) +
- geom_point() +
- stat_smooth(method = "lm", level = .95) +
- scale_x_continuous(labels = function(x) 10 ^ x) +
- labs(x = lab_lines$brain_log, y = lab_lines$sleep_log)
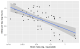
This interval conveys some uncertainty in the estimate of the mean, but this
interval has a frequentist interpretation which can be
unintuitive for this sort of data.
Now, for the point of this post: What’s the Bayesian version of this kind of
visualization? Specifically, we want to illustrate:
- Predictions from a regression model
- Some uncertainty about those predictions
- Raw data used to train the model
The regression line in the classical plot is just one particular line. It’s the
line of best fit that satisfies a least-squares or maximum-likelihood objective.
Our Bayesian model estimates an entire distribution of plausible
regression lines. The first way to visualize our uncertainty is to plot our
own line of best fit along with a sample of other lines from the posterior
distribution of the model.
First, we fit a model RStanARM using weakly informative priors.
- library("rstanarm")
-
- m1 <- stan_glm(
- log_sleep_total ~ log_brainwt,
- family = gaussian(),
- data = msleep,
- prior = normal(0, 3),
- prior_intercept = normal(0, 3))
We now have 4,000 credible regressions lines for our data.
- summary(m1)
- #> stan_glm(formula = log_sleep_total ~ log_brainwt, family = gaussian(),
- #> data = msleep, prior = normal(0, 3), prior_intercept = normal(0,
- #> 3))
- #>
- #> Family: gaussian (identity)
- #> Algorithm: sampling
- #> Posterior sample size: 4000
- #> Observations: 56
- #>
- #> Estimates:
- #> mean sd 2.5% 25% 50% 75% 97.5%
- #> (Intercept) 0.7 0.0 0.6 0.7 0.7 0.8 0.8
- #> log_brainwt -0.1 0.0 -0.2 -0.1 -0.1 -0.1 -0.1
- #> sigma 0.2 0.0 0.1 0.2 0.2 0.2 0.2
- #> mean_PPD 1.0 0.0 0.9 0.9 1.0 1.0 1.0
- #> log-posterior 12.0 1.2 9.0 11.5 12.3 12.9 13.4
- #>
- #> Diagnostics:
- #> mcse Rhat n_eff
- #> (Intercept) 0.0 1.0 3040
- #> log_brainwt 0.0 1.0 3046
- #> sigma 0.0 1.0 2862
- #> mean_PPD 0.0 1.0 3671
- #> log-posterior 0.0 1.0 2159
- #>
- #> For each parameter, mcse is Monte Carlo standard error, n_eff is a crude measure of effective sample size, and Rhat is the potential scale reduction factor on split chains (at convergence Rhat=1).
For models fit by RStanARM, the generic coefficient function coef() returns
the median parameter values.
- coef(m1)
- #> (Intercept) log_brainwt
- #> 0.7354829 -0.1263922
- coef(m1_classical)
- #> (Intercept) log_brainwt
- #> 0.7363492 -0.1264049
We can see that the intercept and slope of the median line is pretty close to
the classical model’s intercept and slope. The median line serves as the “point
estimate” for our model: If we had to summarize the modeled relationship using
just a single number for each parameter, we can use the medians.
One way to visualize our model therefore is to plot our point-estimate line
plus a sample of the other credible lines from our model. First, we create a
data-frame with all 4,000 regression lines.
- # Coercing a model to a data-frame returns data-frame of posterior samples.
- # One row per sample.
- fits <- m1 %>%
- as_data_frame %>%
- rename(intercept = `(Intercept)`) %>%
- select(-sigma)
- fits
- #> # A tibble: 4,000 × 2
- #> intercept log_brainwt
- #> <dbl> <dbl>
- #> 1 0.7529824 -0.1369554
- #> 2 0.7243708 -0.1266290
- #> 3 0.7575502 -0.1171410
- #> 4 0.7855554 -0.1031353
- #> 5 0.6327073 -0.1795992
- #> 6 0.6474521 -0.1714347
- #> 7 0.7512467 -0.1155559
- #> 8 0.7363273 -0.1162038
- #> 9 0.7490401 -0.1276618
- #> 10 0.7238091 -0.1305896
- #> # ... with 3,990 more rows
We now plot the 500 randomly sampled lines from our model with light,
semi-transparent lines.
- # aesthetic controllers
- n_draws <- 500
- alpha_level <- .15
- col_draw <- "grey60"
- col_median <- "#3366FF"
-
- ggplot(msleep) +
- aes(x = log_brainwt, y = log_sleep_total) +
- # Plot a random sample of rows as gray semi-transparent lines
- geom_abline(aes(intercept = intercept, slope = log_brainwt),
- data = sample_n(fits, n_draws), color = col_draw,
- alpha = alpha_level) +
- # Plot the median values in blue
- geom_abline(intercept = median(fits$intercept),
- slope = median(fits$log_brainwt),
- size = 1, color = col_median) +
- geom_point() +
- scale_x_continuous(labels = function(x) 10 ^ x) +
- labs(x = lab_lines$brain_log, y = lab_lines$sleep_log)
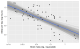
Each of these light lines represents a credible prediction of the mean across
the values of x. As these line pile up on top of each other, they create an
uncertainty band around our line of best fit. More plausible lines are more
likely to be sampled, so these lines overlap and create a uniform color around
the median line. As we move left or right, getting farther away from the mean of
x, the lines start to fan out and we see very faint individual lines for some
of the more extreme (yet still plausible) lines.
The advantage of this plot is that it is a direct visualization of posterior
samples—one line per sample. It provides an estimate for the central tendency
in the data but it also converys uncertainty around that estimate.
This approach has limitations, however. Lines for subgroups require a little
more effort to undo interactions. Also, the regression lines span the whole x
axis which is not appropriate when subgroups only use a portion of the x-axis.
(This limitation is solvable though.) Finally, I haven’t found good defaults
for the aesthetic options: The number of samples, the colors to use, and the
transparency level. One can lose of lots and lots and lots time fiddling with
those knobs!




 雷达卡
雷达卡


















 京公网安备 11010802022788号
京公网安备 11010802022788号







